Abstract
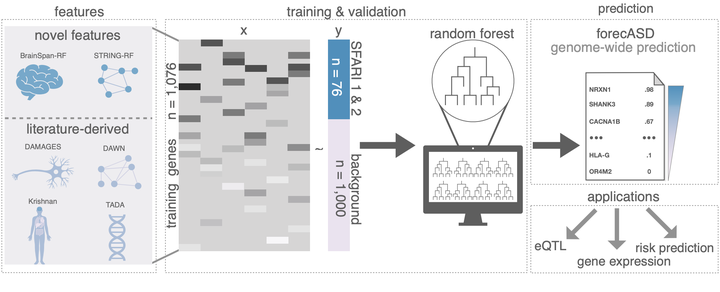
Genes are one of the most powerful windows into the biology of autism, and it has been estimated that perhaps a thousand or more genes may confer risk. However, less than 100 genes are currently viewed as having robust enough evidence to be considered true “autism genes”. Massive genetic studies are underway to produce data to implicate additional genes, but this approach, although necessary, is costly and slow-moving. Here, we approach autism gene discovery as a machine learning problem, rather than a genetic association problem, and use genome-scale data as predictors for identifying further genes that have similar properties in the feature space compared to established autism risk genes. This approach, which we call forecASD, integrates spatiotemporal gene expression, heterogeneous network data, and previous gene-level predictors of autism association to yield a single score that represents each gene’s likelihood of being involved in the etiology of autism. We demonstrate that forecASD has substantially increased sensitivity and specificity compared to previous gene-level predictors of autism association, including genetic-based measures such as TADA. On an independent test set, consisting of newly-released pilot data from the SPARK Genomics Consortium, we show that forecASD best predicts which genes will have an excess of likely gene disrupting (LGD) mutations. Using forecASD results, we show which molecular pathways are currently under-represented in the autism literature and likely represent under-appreciated biological mechanisms of autism. Finally, the larger importance of this work is that by enumerating the genes that are most likely involved in the pathogenesis of autism, we have an opportunity to consider what molecular research in autism might look like in a post-gene discovery era.